Associate Professor, Widener University, PA
Website
Loss of tissue integrity due to injury, physiological or cellular changes is repaired through different mechanisms, such as wound healing, tissue repair or regeneration, and cell turnover. All organisms have the capacity to repair tissue to varying extents after injury, but few animals have true regeneration potential. Promoting the regeneration of post-mitotic cells in a damaged human spinal cord is an important medical goal. My focus is to characterize the changes in gene expression occurring as a result of trauma to the embryonic central nervous system of Drosophila melanogaster, a genetically tractable model organism. Drosophila is a widely used model organism in which genetic, cellular and molecular mechanisms underlying genes implicated in human health and disease have been investigated. Moreover, the availability of powerful genetic tools, a sequenced genome and a comprehensive understanding of developmental processes make Drosophila an ideal system in which to study questions relevant to human health. Total RNA samples were prepared from late stage embryos whose nerve cords were wounded using a fine bore capillary glass needle, and from control unwounded embryos. RNA-seq data generated from these biological samples were analyzed to identify changes in gene expression due solely to injury as opposed to the stress of being handled, and then to determine the gene expression changes occurring post-injury in the embryonic ventral nerve cord versus the extracellular matrix or epithelium.
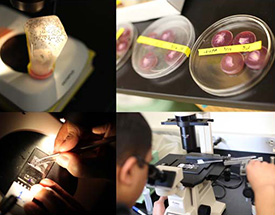
0 –1 hour old embryos from Drosophila melanogaster are collected at room temperature. The embryos are aged for 20 hours at 20oC. The eggshell (chorion) is removed by rolling the eggs on double-sided sticky tape. The dechorionated embryos are aligned on their lateral sides with their ventral aspects exposed on a microscope slide coated with a thin film of glue extracted from preservation-quality sticky tape. Individual embryos are wounded using a fine bore capillary glass needle mounted on a micromanipulator. The embryos are covered with a thin layer of halocarbon oil to prevent desiccation. The procedure is monitored using an inverted compound microscope, first to ensure that the embryos were aligned correctly and are at a stage in development where the nerve cord is fully formed; and second to ensure that the nerve cord was severed by the glass needle. In parallel, samples are generated where embryos are collected, aged, dechorionated and manipulated in a similar manner, but not wounded. Embryos are washed from the slides into scintillation vials using a stream of heptane. Embryo samples are transferred to microfuge tubes and excess heptane is removed. Trizol is added to the semi-dry embryos and the samples are frozen using liquid nitrogen. Dechorionation, line-up, injury (or mock), harvest and freezing steps are completed within one hour. Frozen samples are stored at -80oC. Total RNA was prepared following standard trizol extraction protocol followed by further clean-up using Qiagen RNAeasy Plus kit. Genomic DNA was removed using on-column DNAseI digestion.
RNA-seq dataset to be provided at a later time
Materials are under development.