Chair, Dept of Biological Sciences, Webster University, MO
Isoflurane is a common volatile anesthetic used often in clinical practice; however, its precise molecular effects on cells has not been well described in published literature. We have utilized several model organisms as experimental patients that undergo isoflurane exposure at clinically relevant doses, to assess its cellular effects. One interesting project has shown that Saccharomyces cerevisiae exposed to isoflurane significantly slowed cell growth and reduced cell viability. Several undergraduate senior students have furthered this research. DNA fragmentation is increased (by TUNEL analysis), indicating a possible apoptotic effect. Microarray analysis showed isoflurane exposure reduced the expression of genes for glucose importers HXT2, HXT9, HXT11, and treated cells utilized much less glucose from the culture media.Mitochondrial respiration requires much less glucose than fermentation to sustain life, thus although S. cerevisiae, like many fungi and bacteria, survive largely via fermentation (alternative metabolism), they likely switch to respiration in response to anesthetics. Mutant yeasts (cox9, cox10) lacking genes critical for respiratory metabolism were more susceptible to isoflurane than wild type cells, implying mitochondrial respiration is important for cell survival when challenged with anesthetics. This may have clinical implications when individuals with genetic mitochondrial deficiency diseases must undergo surgery, as their cells will respire less efficiently; there are currently 41 classified mitochondrial disorders in humans (www.umdf.org). Interestingly, yeast cells exposed to these volatile anesthetics begin to undergo a yeast-to-hyphal morphologic switch, a phenomenon similar to that described only once, in the pathogenic fungus Candida albicans.
The specific aim of our project is to perform RNA seq analysis of yeast treated with isoflurane (compared to non treated cells) to directly compare with microarray data. Undergraduate students initiated this project (in 2004) and have conducted all of the subsequent experiments. Microarrays have been analyzed by independent research students as well as in classes. This combination for mining data is very powerful, we wish to broaden it to include next generation sequencing, beginning with RNA seq.
BY4741 yeast were grown to an OD of 0.6 in rich media (YPD). The culture was then split into duplicate treated and non treated samples. Isoflurane was added to the two treatment cultures at a final concentration of 0.1% v/v and yeast were exposed for 30 minutes. The untreated cultures were maintained at the same growth conditions without any addition. Following exposure, RNA was isolated using the hot phenol method. The quality and quantity of RNA was determined spectroscopically and by gel electrophoresis.
RNA-seq dataset to be provided at a later time.
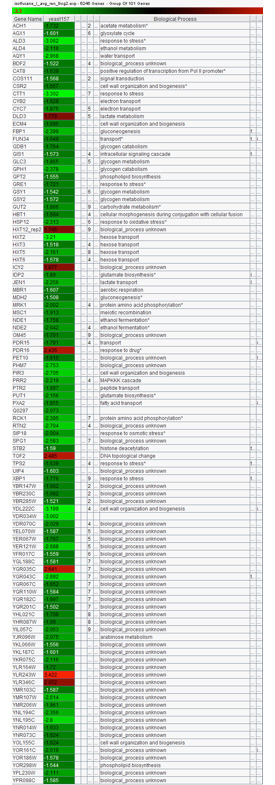
Microarray data analysis from yeast treated for 30 minutes with 0.1% v/v Isoflurane
The Gene Expression course (BIOL 4050) I teach is an upper level elective that is not required for our Biology degrees (highly recommended elective for our Research and Technology emphasis, and required for our BS in Computational Biology degree). In the Gene Expression course, I have used RNA microarrays (yeast and human) obtained through GCAT. First, students analyze previous microarray data (such as the isoflurane yeast microarray). Then, students work in pairs to design a microarray experiment, treat cells, isolate RNA and prepare microarrays. After images are received, the students analyze their data. As a class, we select about 5 genes to follow up with. The students design primers for qRT PCR, and they use qRT PCR to validate microarray findings. I want to include RNA seq analysis in that class. I do realize that it will probably be prohibitively expensive to have that class generate their own RNA seq data. I plan to use the data set obtained in this workshop for next spring’s class to directly compare the results of previous microarray data with RNA seq data.
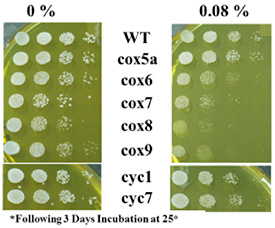
Figure 1: Results of S. cerevisiae cytochrome oxidase mutations grown in the presence of Isoflurane.