Saint Louis University
Website
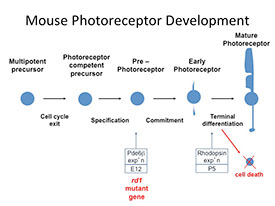
Retinitis pigmentosa (RP) is a genetically heterogeneous group of retinal degenerations leading to night blindness, progressive loss of peripheral visual field, and ultimately may result in complete blindness. Although numerous RP gene defects have been identified, the signaling pathways that trigger cell death are poorly understood. The retinal degeneration1 (rd1) mouse is the best characterized animal model of RP, with early onset degeneration beginning at postnatal day 10 (P10) and rod photoreceptor cell death completed by P21. The mutant gene has been identified, as well as the specific mutation, the protein encoded by that gene (phosphodiesterase-6β), and the endogenous function of that protein in the retina. It plays a critical role in phototransduction, yet it is first expressed in the developing retina at embryonic day 12, approximately two weeks before rhodopsin and other phototransduction genes. Little is known about the molecular cascade of events that link the mutation to cell death in developing rod photoreceptors. The goal of my proposed RNA-seq experiment is to identify differential gene expression in the rd1 retina at P4 and P6. This critical time window represents the earliest identified biochemical and morphological changes in the rd1 retina. Identification of differential gene expression at these time points will provide insights into the molecular and cellular events that precede cell death in this mouse model of retina degeneration in addition to changes that occur during normal retinal development.
Retinal samples were harvested at either postnatal day 4 or 6 from wild type or rd1 mice. All mice were on a C57Bl6/J background. Each sample was comprised of two retinas isolated from one male mouse pup. Replicates were harvested from littermates.
Eight samples have been run for RNA-Seq data: isolated retinas from 2 different time points (postnatal days 4 and 6) for both wild type and rd1 mutant retinas, 2 replicates of each condition. Illumina TrueSeq, stranded, paired ends. Data will be available at a time to be determined.
NCBI SRA links will be provided in Spring 2015.
Developmental Biology is an upper level undergraduate course (taught with an optional lab) with about 20 students, primarily Biology and Neuroscience majors. The course investigates cellular and molecular mechanisms responsible for building multicellular organisms. Students will learn how RNA-Seq can be used to address questions in developmental biology. They will perform analysis on RNA Seq datasets. Students enrolled in the laboratory section will conduct follow-up qPCR reactions to confirm identified genes of interest.